Most recent BDB-Lab papers
BDB-Lab Links
Major Projects
Other Links
Lab Members
Twitter feed
Tweets by BigDataBiologyMethionine tanks: how we almost got fooled by a flawed dataset
by Célio Dias Santos Júnior, Luis Pedro Coelho.
The translation of a given mRNA needs a start codon. This is a particular sequence (typically AUG, although others exist), which indicates the position where translation should start. However, these codons also encode for a methionine residue. Thus, immediately after translation, all proteins have a methionine on their N-terminus. However, it is known that, after translation, there is a process of N-terminal methionine Excision (NME) (Wingfield, 2017). Note that NME is not carried out for all proteins, so that ~20% of the proteins do retain the initial methionine (Frottin et al., 2006).
In evaluating our AMP prediction tool, MACREL, we noticed that some commonly used datasets are inconsistently processed, whereby the negative sequences mostly retain the initial methionine, while the positives sequences do not. This means that the following “classifier” would actually achieve very good results:
if (seq[0] == 'M'):
return 'not-AMP'
else:
return 'AMP'
Obviously, this is nonsense. Like in the urban legend of the neural network that learned to recognize morning and evening tanks in pictures, this dataset was plagued by artefacts.
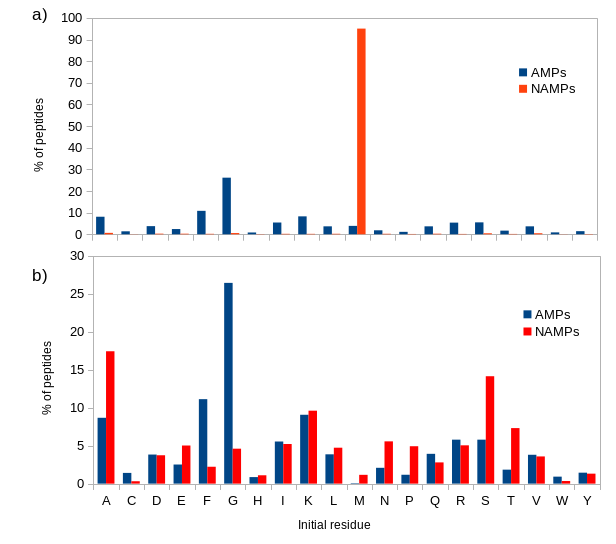
As we do not have a reliable computational method to predict when NME will take place, we opted to always remove the initial Methionine if present, thus avoiding this overfit. This initial methionine removal is implemented in version 0.4 of macrel.
References
- Bhadra P., Yan J., Li J. et al. AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci Rep 8, 1697 (2018). doi: 10.1038/s41598-018-19752-w
- Frottin F., Martinez A., Peynot P., Mitra S., Holz R.C., Giglione C., Meinnel T. The Proteomics of N-terminal Methionine Cleavage. Molecular & Cellular Proteomics 5 (2006), 12, 2336-2349. doi: 10.1074/mcp.M600225-MCP200
- Wingfield P.T. N-terminal methionine processing.Current Protocols in Protein Science 88 (2017), 6141–6143. doi: 10.1002/cpps.29
Copyright (c) 2018–2024. Luis Pedro Coelho and other group members. All rights reserved.